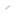Essential tRNA modification stands in the spotlight Modified base determined 40 years ago was a hydrolyzed artifact

The deep understanding of biological processes made possible through genome research and molecular biology is expected to have a significant impact on medical science and pharmaceutical development.
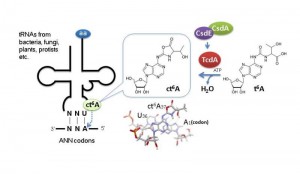
N 6-Threonylcarbamoyladenosine (t6A), a modified nucleotide found in certain tRNAs, has an essential role in translational fidelity. © Tsutomu Suzuki
New analytical data reveal that t6A adopts a cyclic form (ct6A) in cells and has led to the identification of the enzymes that convert t6A to ct6A. ct6A is thought to stabilize codon-anticodon interaction to promote decoding efficiency. Structural modeling suggests that ct6A recognizes the first adenine base of ANN codon at the ribosomal A site.
One of the fundamental tasks carried out by all living organisms is the production of proteins by accurately reading the genetic information (codons) encoded on DNA, initially onto messenger RNA (mRNA). Transfer RNA (tRNA) is an adaptor molecule that deciphers the codons carried on mRNA. tRNAs contain various modified bases which enable them to decipher genetic codes very accurately and efficiently. N 6-threonylcarbamoyldenosine (t6A), identified 40 years ago, is one of the most famous modified bases in tRNAs and is believed in be present in all living organisms. It has been known that t6A is an essential modification and plays crucial roles at multiple stages of protein synthesis. The presence of t6A in cellular tRNAs has been well documented for more than four decades, and research has proceeded on the understanding that t6A was produced in intracellular tRNA.
However, new analytical data by Prof. Suzuki and Project Researcher Kenjyo Miyauchi’s group at the University of Tokyo Graduate School of Engineering has revealed that t6A adopts a cyclic form (named cyclic t6A, ct6A) in bacteria, fungi, plants and protists, demonstrating that t6A is a hydrolyzed artifact of ct6A produced during preparation of tRNA and nucleosides. In fact, they showed evidence that there is no t6A in Escherichia coli, and that an enzyme that converts t6A to ct6A was also identified. Functional data showed that ct6A is involved in promoting decoding efficiency.
This study revealed the true chemical structure of natural RNA modification in living organisms, providing a significant contribution to understanding the molecular mechanism for maintenance of translational fidelity. Since most previous studies on t6A were carried out using E. coli and yeast as model organisms, this discovery will urge scientists to revisit studies related to t6A from the last 40 years.
Paper
Kenjyo Miyauchi, Satoshi Kimura, Tsutomu Suzuki,
“A cyclic form of N 6-threonylcarbamoyladenosine as a widely distributed tRNA hypermodification”,
Nature Chemical Biology Online Edition: 2012/12/16 (Japan time), doi: 10.1038/nchembio.1137.
Article link
Links
Graduate School of Engineering
Department of Chemistry and Biotechnology, Graduate School of Engineering
Suzuki laboratory, Department of Chemistry and Biotechnology, Graduate School of Engineering