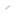Glycoanalysis from a single droplet A new mass spectrometric method for detection of protein glycosylation

Glycosylation is an important and universal post-translational modification for many proteins. Specific changes in protein glycoyslation have been implicated in malignant tumors and therefore development of a simple method to determine glycosylation of limited amounts of individual proteins would be a useful diagnostic tool. MALDI-MS (matrix-assited laser desorption/ionization mass spectrometry) is an indispensable analytical tool for elucidating both the peptide and glycan moieties of glycopeptides. However, it has been difficult to simultaneously analyze glycosylated and nonglycosylated peptides within a proteolytic digest by MS because the ionizaiton efficiency of glycosylated and nonglycosylated peptides is so different. Although several methods have been developed for isolation of glycopeptides, they are time-consuming and give rise to poor yields.
Focusing of hydrophilic peptides by liquid matrix, 3AQ/CHCA © Takuya Shuo
3AQ/CHCA is first spotted onto the MALDI target plate, and then proteolytic whole protein digests containing both glycosylated and nonglycosylated peptides are applied to the top surface of the liquid matrix.
Evaporation of the sample solution allows the droplet to shrink such that the hydrophilic constituents are gradually concentrated in a focused droplet.
A collaborative research team by T. Shuo and Professor M. Seiki (Division of Cancer Cell Research, Institute of Medical Science), and Professor K. Tanaka (Koichi Tanaka Mass Spectrometry Research Lab., Shimadzu Corp.) applied a new technique for MALDI-MS to analyze MT1-MMP (membrane-type 1 matrix metalloproteinase), the protease activity of which is reported to be modulated by glycosylation thereby affecting cancer cell invasion. In this method, a single droplet of proteolytic MT1-MMP digest containing both glycosylated and nonglycosylated peptides is spotted directly onto the liquid matrix 3AQ/CHCA on the MALDI target plate. Gradual evaporation of solvents of the digest on the matrix selectively concentrates hydrophilic glycopeptides at the center, and leaving the hydrophobic components at the periphery. This on-target separation of glycopeptides makes it possible to analyze glycosylated peptides of MT1-MMP in a highly sensitive manner. They demonstrated that MT1-MMP expressed in cancer cells represents a mixture of heterogeneously glycosylated forms. Since cancer cells are reported to have altered glycosylation of proteins, this easy-to-use method for glycopeptide analysis opens up the possibility to identify specific glycosylation patterns of proteins that can be used as new biomarkers for malignant tumors.
Press releasePaper
Takuya Shuo, Naohiko Koshikawa, Daisuke Hoshino, Tomoko Minegishi, Hiroko Ao-Kondo, Masaaki Oyama, Sadanori Sekiya, Shinichi Iwamoto, Koichi Tanaka, Motoharu Seiki,
“Detection of the Heterogeneous O-Glycosylation Profile of MT1-MMP Expressed in Cancer Cells by a Simple MALDI-MS Method”,
PLoS ONE 7(8) (2012) e43751, doi: 10.1371/journal.pone.0043751.
Article link