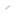Molecular mechanism behind circadian rhythm DNA-binding regions and motifs of CLOCK protein determined

Many aspects of behavior and physiology, including sleep-wake cycles and hormone levels, keep a rhythm with an approximately 24-hour period even under constant conditions. The self-sustaining oscillation mechanism behind these physiological phenomena is called the circadian rhythm, and is generated by a gene transcription/translation-based negative feedback loop. It is known that in this loop at the molecular level, the CLOCK-BMAL1 transcription factor complex, which rhythmically binds to DNA, promotes transcription of various genes, and plays a central role in the circadian rhythm. However, many questions remain about the details of the molecular level control of the circadian rhythm.
© 2014 Hikari Yoshitane.
The DNA-binding of CLOCK protein was investigated using ChIP-Seq analysis using mouse liver. The profiles of sequencing tags at the Dbp gene locus and the heat maps showed rhythmic DNA-binding of CLOCK protein. CLOCK binding motifs were determined by MOCCS analysis from the CLOCK-ChIP Seq data.
The research group of Assistant Professor Hikari Yoshitane, graduate student Hideki Terajima and Professor Yoshitaka Fukada at the Graduate School of Science Department of Biophysics and Biochemistry, collaborating with the research group of graduate student Haruka Ozaki at the Graduate School of Frontier Sciences Department of Computational Biology and Associate Professor Wataru Iwasaki at the Atmosphere and Ocean Research Institute, has identified the CLOCK protein DNA-binding regions throughout the entire mouse genome using next-generation sequencers. Collectively, dynamic circadian outputs are orchestrated by CLOCK-mediated direct transcription in combination with indirect regulation, such as transcription by CLOCK-regulated transcription factors and gene silencing by CLOCK-regulated miRNAs. Furthermore, the research group developed a new bioinformatics method MOCCS (motif centrality analysis of ChIP-Seq; Method for determination of DNA nucleotide sequences recognized by DNA-binding proteins from ChIP-Seq data) and used this method to identify the DNA nucleotide sequences of the binding sites of the CLOCK transcription factor.
The newly-developed MOCCS motif-finding algorithm could be adopted by a wide range of ChIP-sequencing studies for the genome-wide identification of protein-DNA binding regions.
Paper
Hikari Yoshitane, Haruka Ozaki, Hideki Terajima, Ngoc-Hien Du, Yutaka Suzuki, Taihei Fujimori, Naoki Kosaka, Shigeki Shimba, Sumio Sugano, Toshihisa Takagi, Wataru Iwasaki, and Yoshitaka Fukada,
“CLOCK-controlled polyphonic regulations of circadian rhythms through canonical and non-canonical E-boxes”,
Molecular and Cellular Biology 2014, May, 34(10), 1776-1787, doi: 10.1128/MCB.01465-13.
Article link
Links
Department of Biological Sciences, Graduate School of Science (Japanese)
Fukada Laboratory, Department of Biological Sciences, Graduate School of Science