Newton's cradle proton relay in cellulase Visualizing protein versatility and ingenuity by neutron diffraction

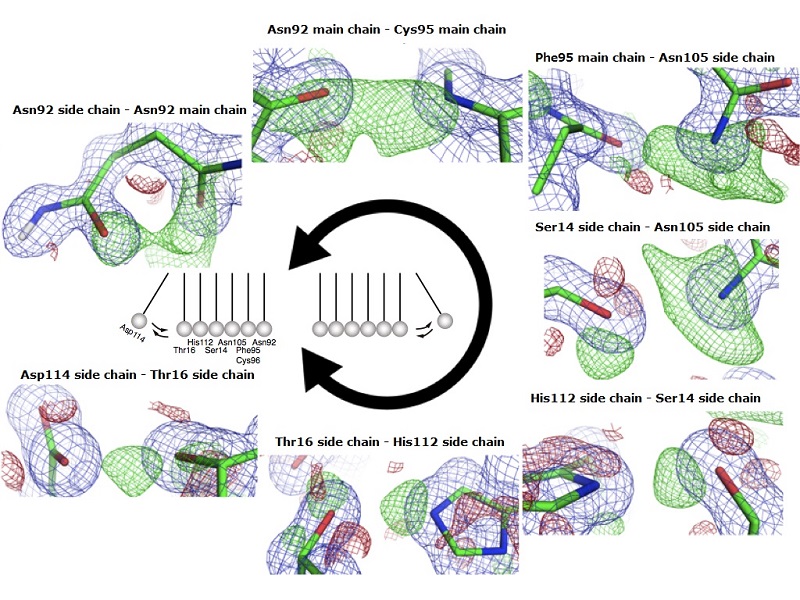
Newton’s cradle-like proton relay network around important catalytic amino acids in fungal cellulase PcCel45A
A proton relay pathway is formed between two important catalytic residues, the imidic acid of Asparagine at the 92nd position, through peptide bonds of Cysteine at 96 and Phenylalanine at 95, the imidic acid of Asparagine at 105, side chains of Serine at 14, Histidine at 112, Threonine at 16, and finally to Asparagine at 114. The proton moves back and forth along this route (green mesh) like energy in a Newton’s cradle.
© 2015 Kiyoihko Igarashi.
Researchers at the University of Tokyo have demonstrated that during cellulose catalysis by cellulase produced by wood-degrading fungi, protons are transferred in a manner similar to energy in a Newton’s cradle. This research shows the importance of neutron diffraction studies to visualize the versatility and ingenuity of proteins, overlooked in former structural studies.
Cellulose is one of the main components of the plant cell wall and the most abundant form of biomass on Earth. The looming threat of exhaustion of fossil fuel resources means that it is becoming important to develop technologies to degrade and convert biomasses such as wood and grass to a variety of materials.
In the present research, the research group of Associate Professor Kiyohiko Igarashi at the University of Tokyo Graduate School of Agricultural and Life Sciences Department of Biomaterial Sciences successfully solved the neutron structure of large-volume cellulase crystals (6 mm3) obtained from a wood-degrading fungus (PcCel45A) using the Ibaraki biological crystal diffractometer (iBIX) at the Materials and Life Science Experimental Facility (MLF) in the Japan Proton Accelerator Research Complex (J-PARC). It was revealed that the amino acid Asparagine in the “imidic acid form” is catalytically important for the function of this enzyme. Moreover, the protons are transferred back and forth via the Asparagine residue, like energy in a Newton’s cradle, and this movement is the key for the repetition of the reaction by the enzyme.
“This research suggests that the same phenomenon may occur in a variety of other proteins,” says Associate Professor Igarashi. He continues, “As a result, this innovative discovery will lead not only to a better understanding of the reaction mechanism by which enzymes degrade biomass, but also find applications in areas such as computer-aided drug design.”
This research was carried out in collaboration with Ibaraki University, Japan Aerospace Exploration Agency (JAXA), University of the Ryukyus, Confocal Science Inc., Maruwa Foods and Biosciences Inc., University of Hyogo, and Ibaraki Prefecture.
Press release (Japanese)
Paper
, ""Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography", Science Advances Online Edition: 2015/8/22 (Japan time), doi: 10.1126/sciadv.1500263.
Article link (Publication)
Links
Graduate School of Agricultural and Life Sciences
Department of Biomaterial Sciences, Graduate School of Agricultural and Life Sciences






