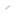Similarities in moth pheromone synthesis and human stress/appetite control Ligand recognition site of receptor controlling moth pheromone biosynthesis identified

The successful propagation of many moth species is dependent on the female’s ability to attract conspecific (same-species) males via species-specific sex pheromones. In most Lepidoptera, pheromone biosynthesis is regulated by a C-terminally amidated 33-amino acid neuropeptide termed pheromone biosynthesis-activating neuropeptide (PBAN) that originates from a part of the central nervous system within the head. The sex pheromone is produced when PBAN binds to the PBAN receptor (PBANR) in the pheromone glands in the posterior section of the abdomen. The binding mode of PBAN to PBANR and the amino acid residues important for PBANR function, however, have not been as well defined.
© 2014 Masaru Tanokura, Koji Nagata.
Location of functionally important residues in PBANR. The important residues identified by Ala substitution for (A) PBAN binding, and (B) Ca2+ mobilization are colored and labeled in the homology-based model of PBANR. (C) The docking model of the minimal active fragment of PBAN (C5; F-S-P-R-L-NH2) and PBANR. The bound ligand (C5) is depicted in the space-filling model. The seven transmembrane helices (TMs) in PBANR are numbered as TM1-TM7 from the N-terminus.
In the current study, the group of Professor Masaru Tanokura and Associate Professor Koji Nagata at the Graduate School of Agricultural and Life Sciences, UTokyo, used point mutation analysis to identify the important amino acid residues of PBANR for (1) its translocation to the plasma membrane, (2) its binding to PBAN, and (3) Ca2+ mobilization induced by PBAN binding to PBANR. Additionally, they used computer simulations to construct a three-dimensional model of the PBAN-PBANR complex that fitted with the experimentally-derived data. They also constructed a three-dimensional model of human neuromedin U (NMU) and its receptor (NMUR1), an evolutionarily-related (orthologous) ligand-receptor pair to PBAN-PBANR, and suggested that both PBAN and NMU dock with their receptors in a similar fashion.
The location of the amino acid residues underlying PBANR function identified in this study corresponds closely with that of G protein-coupled receptors (GPCR) for which crystallographic structures are already known, such as A2A adenosine receptor and &beta2 adrenergic receptor, but a few critical residues specific to PBANR and NMUR1 were also identified. These results will be useful not only in the development of pesticides that block the PBAN?PBANR signaling, but also provide new information concerning the interaction mechanism of NMU-NMUR1, which is involved in the regulation of human appetite, stress and pain.
Press release (Japanese)
Paper
, "Identification of Functionally Important Residues of the Silkmoth Pheromone Biosynthesis-activating Neuropeptide Receptor, an Insect Ortholog of the Vertebrate Neuromedin U Receptor", The Journal of Biological Chemistry 289 (2014): 19150-19163, doi: 10.1074/jbc.M113.488999.
Article link (Publication, UTokyo Repository)
Links
Graduate School of Agricultural and Life Sciences
Department of Applied Biological Chemistry, Graduate School of Agricultural and Life Sciences