Toward prevention of gastric cancer 3D structure of H. pylori CagA Oncoprotein elucidated

The
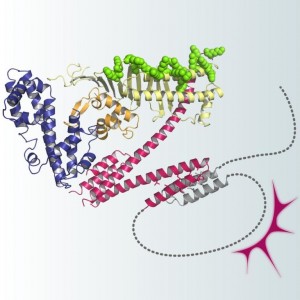
The CagA body comprises three discrete domains (I-III), in which domains II and III constitute a N-shaped core structure. Domain II also contains a basic patch that interacts with phosphatidylserine, an acidic phospholipid concentrated to the inner face of the plasma membrane. The disordered CagA tail loops back onto the structured CagA body to form a lariat-like structure. The intramolecular loop formation stabilizes the interaction of CagA with host proteins such as SHP2 and PAR1 and thereby enhances the oncogenic potential of H. pylori CagA.
The CagA protein of Helicobacter pylori plays a key role in the development of gastric cancer, the second-leading cause of the cancer-related death worldwide. After delivery into host gastric epithelial cells via type IV secretion, CagA promotes gastric carcinogenesis by promiscuously interacting with a variety of host signaling proteins. However, nothing is known with regard to the molecular structure of CagA, and the structural basis underlying the oncogenic action of CagA remains totally unknown. In the present work, a research group led by Prof. Masanori Hatakeyama at the Graduate School of Medicine, the University of Tokyo, and Dr. Toshiya Senda at the National Institute of Advanced Industrial Science and Technology (AIST) succeeded in elucidating the entire 3D structure of the H. pylori CagA oncoprotein by combining X-ray crystallography and NMR data. They found that CagA comprises a folded N-terminal region (CagA body) and an intrinsically disordered C-terminal region (CagA tail). They also found that the disordered CagA tail loops back onto the structured N-terminal body to form a lariat and that the loop formation enhances the oncogenic potential of H. pylori CagA by stabilizing CagA-host protein interaction. This is the first comprehensive study of the CagA structure. The work substantially advances our understanding of H. pylori-mediated gastric carcinogenesis and opens the way for the development of new strategies toward the prevention and control of gastric cancer.
Press release (Japanese)
Paper
Takeru Hayashi, Miki Senda, Hiroko Morohashi, Hideaki Higashi, Masafumi Horio, Yui Kashiba, Lisa Nagase, Daisuke Sasaya, Tomohiro Shimizu, Nagarajan Venugopalan, Hiroyuki Kumeta, Nobuo N. Noda, Fuyuhiko Inagaki, Toshiya Senda, Masanori Hatakeyama,
“Tertiary Structure-Function Analysis Reveals the Pathogenic Signaling Potentiation Mechanism of Helicobacter pylori Oncogenic Effector CagA”,
Cell Host & Microbe Online Edition: 2012/7/19 (Japan time), doi: 10.1016/j.chom.2012.05.010.
Article link