How plants can tell friend from foe Nucleotide “tail” in RNA blocks triggering immune response Announcement

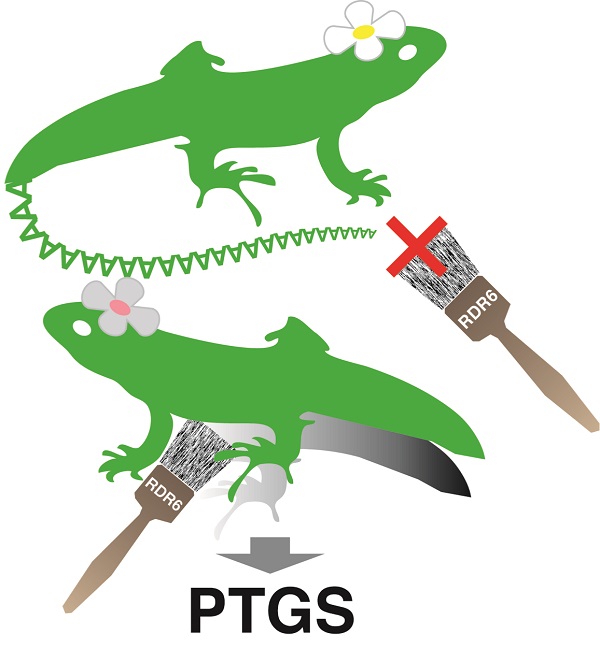
Immune mechanism in plants for distinguishing normal RNAs from abnormal RNAs, to be broken down by post-transcriptional gene silencing (PTGS)
At top, the 3´ poly(A) tail functions as a marker for a normal messenger RNA (lizard) to foil attack by RDR6 (paintbrush) and avoid elimination by PTGS; in contrast, RDR6 converts the aberrant poly(A) tail-less RNA, at bottom, into its double-stranded form, calling on PTGS to destroy it.
© 2017 Kyungmin Baeg, Hiro-oki Iwakawa, Yukihide Tomari.
The plant’s immune system can recognize whether a piece of RNA is an invader or not based on whether the RNA has a threaded bead-like structure at the end, say University of Tokyo researchers. Their finding provides an answer to the quarter-century-old question of why RNAs belonging to the plant escape its self-defense mechanism, paving the way for future biotechnological techniques to modify crops.
Our immune system protects us from diseases and infections by fighting off viruses and other foreign substances. Plants also have a mechanism that fends off invaders, called post-transcriptional gene silencing, which attacks foreign RNA by distinguishing them from non-foreign ones, but does not fight against RNAs originating in the plant.
“While previous research has suggested that a long chain of adenosine nucleotides at the end of the RNA, called the poly(A) tail, and/or a distinctive structure at the beginning of the RNA, the cap structure, could be the flags that dictate whether or not to trigger the plant immune system, no research until now was able to provide definitive evidence,” explains Assistant Professor Hiro-oki Iwakawa of the University of Tokyo’s Institute of Molecular and Cellular Biosciences, co-author of the study to appear in Nature Plants.
“Classical biochemistry techniques are one of our lab’s strengths. With classical biochemistry, we can replicate biological processes in test tubes, and identify and describe the molecules involved. And this is exactly what we have done,” says Professor Yukihide Tomari, also of the University of Tokyo’s Institute of Molecular and Cellular Biosciences.
Tomari and lab members Iwakawa and PhD student Kyungmin Baeg produced an RNA-dependent RNA polymerase 6 (RDR6) protein, a key enzyme in initiating the defense mechanism, from Arabidopsis thaliana, a small flowering plant, and observed how RDR6 behaves when mixed with foreign RNA containing a poly(A) tail of varying length.
The research group found that if the RNAs contained a poly(A) tail at their ends, the immune system did not consider the RNA an invader, while RNAs lacking the tail were perceived as foreign, triggering a defense response.
“Initially, we hypothesized that multiple proteins would be involved in discriminating the two RNAs,” says Baeg. He continues, “To our surprise, it turned out that RDR6 itself can discriminate the presence of the poly(A) tail and this single-enzyme system is much simpler and more elegant than we initially thought.”
Papers
Kyungmin Baeg, Hiro-oki Iwakawa, and Yukihide Tomari, "The poly(A) tail blocks RDR6 from converting self mRNAs into substrates for gene silencing," Nature Plants Online Edition: March 21, 2017, doi:10.1038/nplants.2017.36.
Link (Publication )
)
Related links
- Lab of RNA Function (Tomari Lab), Institute for Quantitative Biosciences

- Institute for Quantitative Biosciences

- Graduate School of Frontier Sciences

- Department of Computational Biology and Medical Sciences, Graduate School of Frontier Sciences

- Grants-in-Aid for Scientific Research on Innovative Areas “Non-coding RNA neo-taxonomy”

- Grants-in-Aid for Scientific Research on Innovative Areas “Nascent-chain Biology” (Japanese)