How an important enzyme used in drug production recognizes its substrate Increased use of enzymes producing chiral amines

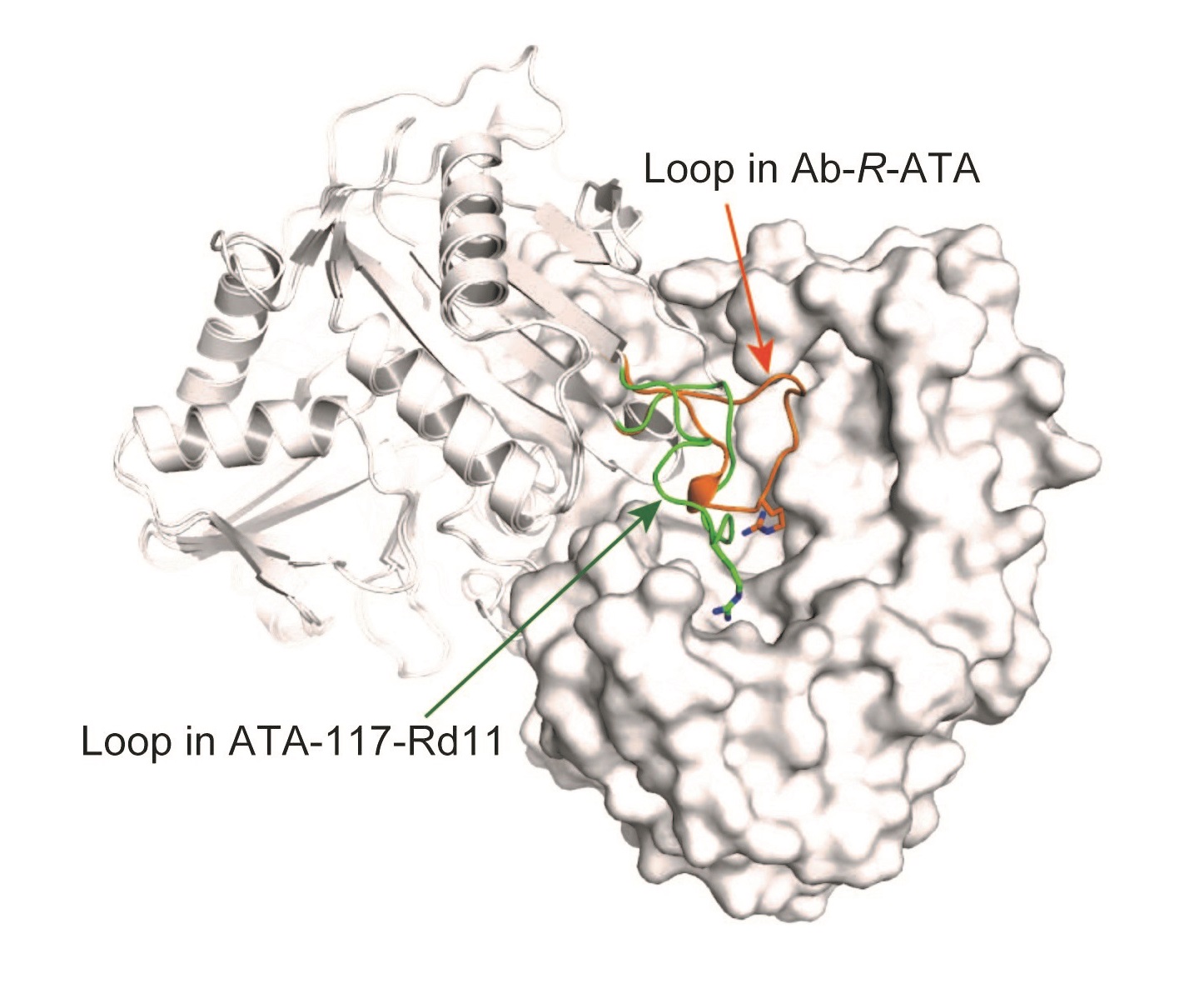
Comparison of loop conformation of Ab-R-ATA and artificially produced ATA-117-Rd11
Superimposed dimeric structures of Ab-R-ATA and ATA-117-Rd11. One protomer of the dimer is shown in surface representation (right) and the other is drawn as a ribbon diagram (left). The loops near the active sites of Ab-R-ATA and ATA-117-Rd11 are highlighted in orange and green, respectively, clearly showing the difference in structure.
© 2015 Masaru Tanokura.
A research group at the University of Tokyo has demonstrated the mechanism by which an enzyme used in industrial production of an important drug for type-2 diabetes recognizes its target. This finding will further design of novel biocatalysts for industrial production of chiral amine compounds, crucial building blocks in the synthesis of some pharmaceutical drugs.
Pure chiral amines are crucial building blocks in the synthesis of some pharmaceutical drugs, agrochemicals and other chemical compounds. Amine transaminases (ATAs) are the focus of increasing attention as useful tools for efficient chiral-amine production. In particular, R-stereospecific ATA (R-ATA), a type of ATA that produces only R-chiral amines, has great commercial value. In 2010 two US pharmaceutical companies created a variant of an R-ATA allowing industrial-scale synthesis of sitagliptin, a medicine for type-2 diabetes. The improved enzyme gave a 53% increase in daily production volume and a 19% reduction in total cost, as compared to the chemical approach conventionally used to synthesize sitagliptin.
However, how the industrially-important enzyme R-ATA recognized its substrate and other details remained unclear.
The research group of Professor Masaru Tanokura at the Graduate School of Agricultural and Life Sciences of the University of Tokyo, in collaboration with Biotechnology and Development Laboratories of Kaneka Corporation, determined the crystal structures of R-ATA from a bacteria that lives in soil and other materials, Arthrobacter sp. KNK168 (Ab-R-RTA), and demonstrated the mechanism of the substrate recognition and substrate specificity of R-ATAs. One unique characteristic of ATAs is that they recognize two different substrates in the same active site, and it was found that the active-site residue arginine (Arg138) functions on both substrates. Moreover, the structures of an artificial R-ATA and the G136F mutant of Ab-R-ATA (created by substituting the 136th base glycine for phenylalanine) revealed that a loop near the catalytic site was the target site that allowed the change in substrate specificity.
“These findings should lead to the rational design of R-ATAs based on this structural information to develop novel biocatalysts useful for the production of a diverse range of chiral amine compounds. This would accelerate the industrial uses of chiral-amine synthesis using R-ATAs,” says Professor Tanokura.
Paper
, "A new target region for changing the substrate specificity of amine transaminases", Scientific Reports: 2015/06/01 (Japan time), doi: 10.1038/srep10753.
Article link (Publication, UTokyo Repository)
Links
Graduate School of Agricultural and Life Sciences
Department of Applied Biological Chemistry, Graduate School of Agricultural and Life Sciences






