Winding DNA by molecular reel A novel method of manipulating DNA’s curvature

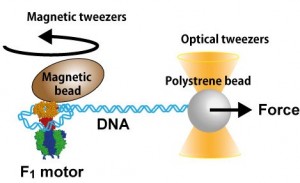
DNA, a long string-like polymer encoding the genetic information of life, is packaged in cells very compactly. Keeping DNA in compact, tightly bent conformation in the cell is important for storage of genetic information and regulation of gene expression. However, the force and energy required for bending DNA tightly with curvature radii of a few nanometers have not been measured directly, only estimated from the relaxed conformation of DNA.
Prof. Hiroyuki Noji’s group at the Department of Applied Chemistry in the University of Tokyo’s Graduate School of Engineering has developed a method to address this issue. By using the F1-ATPase rotary motor protein and single-molecule manipulation techniques, the researchers were able to wind a single DNA molecule into a loop with diameter of 8.5 to 20 nm and directly measure the force required to bend DNA tightly.
This finding by Prof. Noji’s group contributes to restrict the models of DNA-protein interaction in terms of the elastic energy of DNA. This novel approach will also be a useful tool to reveal the mechanism of gene regulation in the cell and contribute to the designing of future DNA-based nano-structures.
Paper
Huijuan You, Ryota Iino, Rikiya Watanabe, Hiroyuki Noji,
“Winding single-molecule double-stranded DNA on a nanometer-sized reel”,
Nucleic Acids Research Online Edition: 2012/07/05 (Japan time), doi: 10.1093/nar/gks65.
Article link
Links
Graduate School of Engineering
Department of Applied Chemistry
Noji Laboratory (Japanese)