Reconstruction of molecular network from cellular big data Global landscape of metabolic regulation by insulin

The cell comprises molecules with a variety of physical properties: DNA, RNA, proteins, and metabolites. A group of molecules with similar physical properties is called an ‘omic layer’. A variety of cellular functions are realized by intracellular molecular networks that connect multiple omic layers. However, the molecular networks between omic layers remain largely unknown.
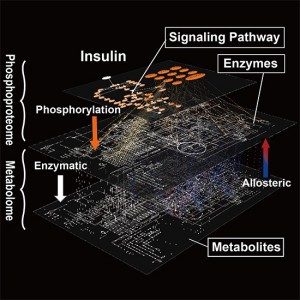
Graphical abstract from Yugi et al. (2014) Cell Reports (CC BY 3.0)
A global molecular network of metabolic regulation by insulin underlying across the protein phosphorylation layer and metabolite layer. The trans-omic analysis, a methodology developed in this study, enabled the reconstruction of the entire landscape of the global network of insulin regulation of metabolism that could have been only guessed at from a patchwork of partial knowledge.
Katsuyuki Yugi, Hiroyuki Kubota, and Shinya Kuroda (Department of Biological Sciences, Graduate School of Science, University of Tokyo) have developed a methodology named ‘trans-omic’ analysis that enables researchers to reconstruct the entire molecular network across two omic layers that include protein phosphorylation and metabolites. This project was performed in collaboration with Tomoyoshi Soga and Kazutaka Ikeda (Institute for Advanced Biosciences, Keio University), Keiichi I. Nakayama and Masaki Matsumoto (Medical Institute of Bioregulation, Kyushu University), Hiroaki Miki and Yosuke Funato (Research Institute for Microbial Diseases, Osaka University). The research group applied trans-omic analysis to metabolic regulation by insulin, dosing rat liver cells with insulin to gather information (big data) about the relationship between protein phosphorylation and metabolites. This big data analysis described the global landscape of metabolic regulation by insulin that could have been only guessed at from a patchwork of partial knowledge.
Further detailed analysis of the reconstructed network of metabolic regulation by insulin may potentially provide novel mechanisms of controlling blood glucose level. In fact, the research group discovered an important blood-sugar regulation network that functions only in the liver. Trans-omic analysis, which is established in this study, is a methodology that is applicable to reconstruction of molecular networks underlying numerous biological functions, not only the metabolic regulation by insulin. Identification of intracellular molecular networks allows not only elucidating the mechanisms of biological functions and diseases but also controlling them. Thus, future developments of trans-omic analysis may facilitate reconstruction of the molecular networks specifically activated in disease model mice, which could serve as a navigation system to search for novel biomarkers and drug targets.
Press release (Japanese)
Paper
Katsuyuki Yugi, Hiroyuki Kubota, Yu Toyoshima, Rei Noguchi, Kentaro Kawata, Yasunori Komori, Shinsuke Uda, Katsuyuki Kunida, Yoko Tomizawa, Yosuke Funato, Hiroaki Miki, Masaki Matsumoto, Keiichi I. Nakayama, Kasumi Kashikura, Keiko Endo, Kazutaka Ikeda,Tomoyoshi Soga and Shinya Kuroda,
“Reconstruction of insulin signal flow from phosphoproteome and metabolome data”,
Cell Reports Online Edition: 2014/August/15 (Japan time), doi: 10.1016/j.celrep.2014.07.021.
Article link (publication, UTokyo Repository)
Links
Department of Biological Sciences, Graduate School of Science
Kuroda Laboratory, Department of Biological Sciences, Graduate School of Science
Bioinformatics and Systems Biology, Graduate School of Science (Japanese)






