Unexpected modification to support maturation of transfer RNA Pre-tRNAs in budding yeast stabilized by the cap modifications

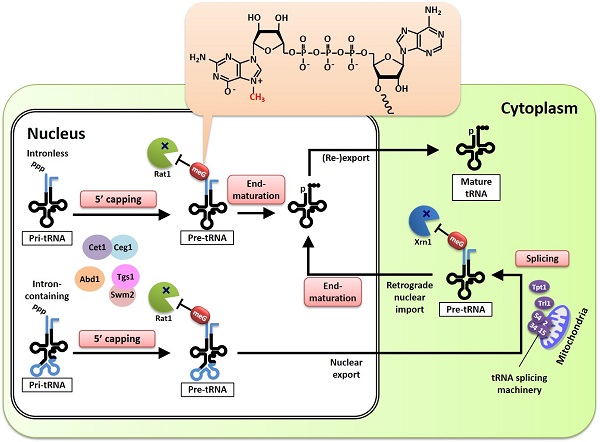
Pre-tRNAs are protected from 5’ exonucleases by 5’ capping during maturation
Pre-tRNAs with 5’ leader sequences are capped at the 5’ termini by canonical enzymes (Ceg1, Cet1, Abd1, Tgs1 and Swm2), protecting them from 5’ exonucleases (Rat1 and Xrn1). The cap’s chemical structure—7-methylguanosine (m7G)—is depicted in the orange-shaded balloon on top.
© 2016 Tsutomu Suzuki.
Researchers at the University of Tokyo discovered cap structures in a certain population of tRNA precursors and showed these modifications support tRNA maturation. The finding upends conventional understanding of RNA cap structures, and suggests that a new mechanism involving RNA capping that serves to regulate gene expression may exist.
Transfer (t)RNA, the key player in deciphering the genetic code, is synthesized as a primary transcript with extra sequences called 5’ leader and 3’ trailer sequences at both its termini. Following transcription, tRNA maturates through removal of these extra sequences and various chemical modifications to acquire its primary function. However, the precise timing and order in which the simple tRNA precursor molecule undergoes modifications to become more complex has not been thoroughly understood.
The University of Tokyo research group led by Assistant Professor Takayuki Ohira and Professor Tsutomu Suzuki of the Department of Chemistry and Biotechnology at the Graduate School of Engineering employed their original methods for RNA isolation and highly sensitive mass spectrometry to analyze the precise chemical structure of tRNA precursors (pre-tRNAs) at various stages in budding yeast.
As a result, the researchers discovered methylguanosine cap structures at the 5’ termini of pre-tRNAs with unprocessed 5’ leader sequence naming the phenomenon “pre-tRNA capping.” In general, the cap structure is found in RNAs transcribed by a particular enzyme, RNA polymerase II; however, as tRNA is generated by a different enzyme, RNA polymerase III, the current finding upends a generally accepted principle in molecular biology. Furthermore, curiously enough, the level of pre-tRNA decreased significantly when cap formation was blocked; in addition, when the activity of an enzyme that degrades tRNA from the 5’ end, 5’ exonuclease, was inhibited, the level of pre-tRNA increased. These results suggest that the cap structures protect the pre-tRNA from being broken down. For tRNA maturation, pre-tRNA needs to move dynamically between cellular compartments. The cap structures likely act as a shield to protect pre-tRNA in the maturation process. This study for the first time reported the phenomenon of pre-tRNA capping, and shed light on some of its functions in tRNA maturation.
“’Transfer RNA precursors are capped!’ With this exciting study, which began with a chance discovery, I was again reminded of the complexity and curiosity of life,” says Ohira. He continues, “I think our findings provided only one aspect of the functional roles of pre-tRNA capping, but I believe that further studies will provide deeper understanding of pre-tRNA capping and further reveal its biological significance.”
Paper
, "Precursors of tRNA are stabilized by methylguanosine cap structures", Nature Chemical Biology Online Edition: 2016/06/27 (Japan time), doi: 10.1038/nchembio.2117.
Article link (Publication)
Links
Graduate School of Engineering
Department of Chemistry and Biotechnology, Graduate School of Engineering
Suzuki Laboratory, Department of Chemistry and Biotechnology, Graduate School of Engineering






