Regulators of metabolism, diabetes identified with new technique Kinetic trans-omic approach may be used to understand other metabolic diseases
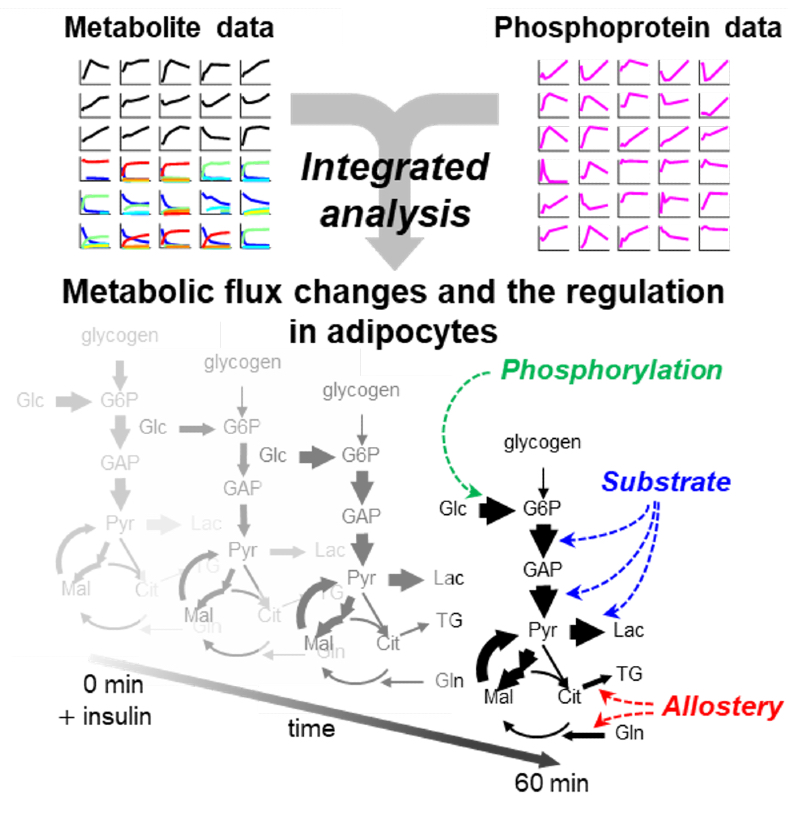
Regulation of glucose metabolism in insulin-stimulated mouse adipocytes was revealed by integrated analysis with comprehensive metabolite and phophoprotein data. © 2020 Shinya Kuroda, all rights reserved
Researchers used a new approach to understand how insulin, the hormone connected to diabetes, changes metabolism. Using lab-grown mouse fat cells, they identified 10 reactions that are regulated by a small number of enzymes.
“It was surprising that regulation of only a few key enzymes was sufficient to induce changes in overall insulin-induced glucose metabolism,” said Professor Shinya Kuroda, M.D., Ph.D., who led the research project at the University of Tokyo.
Kuroda is interested in how cells’ inner communication pathways control biological systems. The system of cellular metabolism includes many communication pathways that regulate nutrient storage and energy production, all influenced or controlled by insulin.
The research team previously identified cell signaling pathways related to diabetes using trans-omics, a broad-scope approach that combines information from databases about biochemical reactions with lab experiments focused on genes (genomics), proteins (proteomics) and metabolites (metabolomics). Now the researchers have added another layer of information by developing a mathematical model of kinetics, or the rates of chemical reactions. Specifically, they used this kinetic trans-omics approach to study the transfer of tiny molecules called phosphate groups onto proteins and the flow of glucose through metabolism.
Tracking the movement of these molecules in great detail and over time allows researchers to understand the relative contributions that individual regulation mechanisms make to the complex task of maintaining metabolism. The mouse fat cells used in this study were actively stimulated during the experiments by adding insulin to their growth liquid.
The team’s experiments revealed that most of the metabolic reaction pathways identified are regulated by the amounts of the chemical ingredients or products of the reactions, rather than by changes to the enzymes that act on those ingredients and products. In particular, much of insulin’s effect on cells seemed to occur by changing the activity of one cell-surface protein, called Glut4, which pulls glucose into fat cells.
The kinetic trans-omics technique can also be applied to a variety of other cells and organs to understand diseases beyond diabetes.
“We expect our approach to be a fundamental technology for understanding and treating the pathology of metabolic diseases, such as Type 2 diabetes,” said Kuroda.
Collaborators at the University of Sydney (Australia), RIKEN Center for Integrative Medical Sciences and Keio University also contributed to this research. This research is a peer-reviewed experimental study using mouse cells published in iScience, a Cell Press journal.
Papers
Satoshi Ohno, Lake-Ee Quek, James R. Krycer, Katsuyuki Yugi, Akiyoshi Hirayama, Satsuki Ikeda, Futaba Shoji, Kumi Suzuki, Tomoyoshi Soga, David E. James, and Shinya Kuroda, "Kinetic trans-omic analysis reveals key regulatory mechanisms for insulin-regulated glucose metabolism in adipocytes," iScience: August 20, 2020, doi:10.1016/j.isci.2020.101479.
Link (Publication )
)





