10,000 leagues under the cell Researchers give nautical name to newly characterized protein important for gene regulation by microRNAs Research news

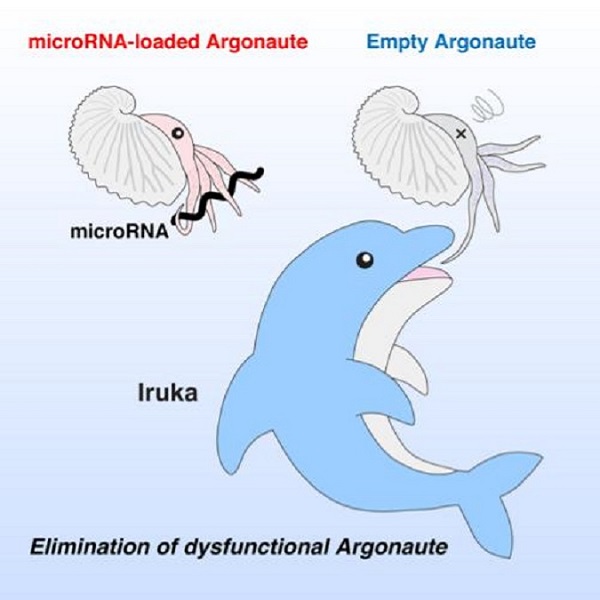
Researchers have identified an elegant mechanism that ensures the function of microRNAs, tiny molecules that control gene activity inside plants and animals, including humans. MicroRNAs cannot function alone; they always need to work together with their partner protein called Argonaute. A research team in the Institute for Quantitative Biosciences at the University of Tokyo discovered a novel protein that triggers degradation of microRNA-free "empty" Argonaute inside cells.
"It has been known that Argonaute proteins are selectively degraded in their empty state, and that this degradation of empty Argonaute is evolutionarily conserved, which implies that this process is important for microRNA function. However, the degradation mechanism and biological significance were unclear. We were curious about this phenomenon," said Professor Yukihide Tomari who supervised the research.
Researchers within the Laboratory of RNA Function, which is led by Tomari, set out to understand how cells can distinguish between empty and microRNA-loaded Argonaute, and why empty Argonaute is degraded.
The research team used fruit fly (Drosophila) cells. Mammals load microRNAs into four different Argonaute proteins (Argonaute1–4), but fruit flies generally sort microRNAs into a single Argonaute (Argonaute1), simplifying analysis of the microRNA pathway.
To identify the protein responsible for degrading empty Argonaute, researchers first purified the binding proteins of empty or microRNA-loaded Argonaute1 and then determined their identities. The research team completed the process of identification using a technique called liquid chromatography tandem mass spectrometry (LC-MS/MS), which can identify the proteins by measuring the precise mass of protein fragments.
"We were excited when we discovered the protein responsible for degrading empty Argonaute. At the same time, we realized that this protein had remained uncharacterized in flies and only had a serial number (CG11982) with no meaningful name," said Research Associate Hotaka Kobayashi, first author of the recent research publication.
Argonaute proteins were originally identified in a plant species called Arabidopsis commonly used in genetic experiments. Arabidopsis plants with a mutation in the argonaute1 gene have tubular, curled leaves that reminded the scientists who discovered them of the tentacles of Argonauta octopuses, also known as paper nautiluses.
To keep with the nautical theme, the UTokyo research team chose to remove the serial number and name the protein Iruka, the Japanese word for dolphin, because dolphins are reported to eat a lot of Argonauta octopuses.
"Discussing the candidate names was a lot of fun," said Kobayashi.
Iruka preferentially binds empty Argonaute and attaches a "destruction mark" called ubiquitin to trigger its degradation. This action of Iruka is important to eliminate dysfunctional Argonaute and ensure effective microRNA-mediated gene silencing.
"Argonaute and microRNAs influence a wide variety of biological processes and diseases including cancer. The newly discovered mechanism of Iruka to ensure microRNA-mediated gene silencing may contribute to understanding microRNA functions and related diseases," said Tomari.
Funding was provided by the Japanese Ministry of Education, Culture, Sports, Science and Technology (MEXT) and the Japan Society for the Promotion of Science (JSPS).
Papers
Hotaka Kobayashi, Keisuke Shoji, Kaori Kiyokawa, Lumi Negishi, Yukihide Tomari, "Iruka eliminates dysfunctional Argonaute by selective ubiquitination of its empty state," Molecular Cell: November 29, 2018, doi:10.1016/j.molcel.2018.10.033.
Link (Publication )
)
Related links
- Institute for Quantitative Biosciences

- Department of Computational Biology and Medical Sciences, Graduate School of Frontier Sciences

- Laboratory of RNA Function (Tomari Lab)

- Interview with Professor Yukihide Tomari






