Beyond finding a gene Same repeated stretch of DNA identified in three neurodegenerative diseases Research news

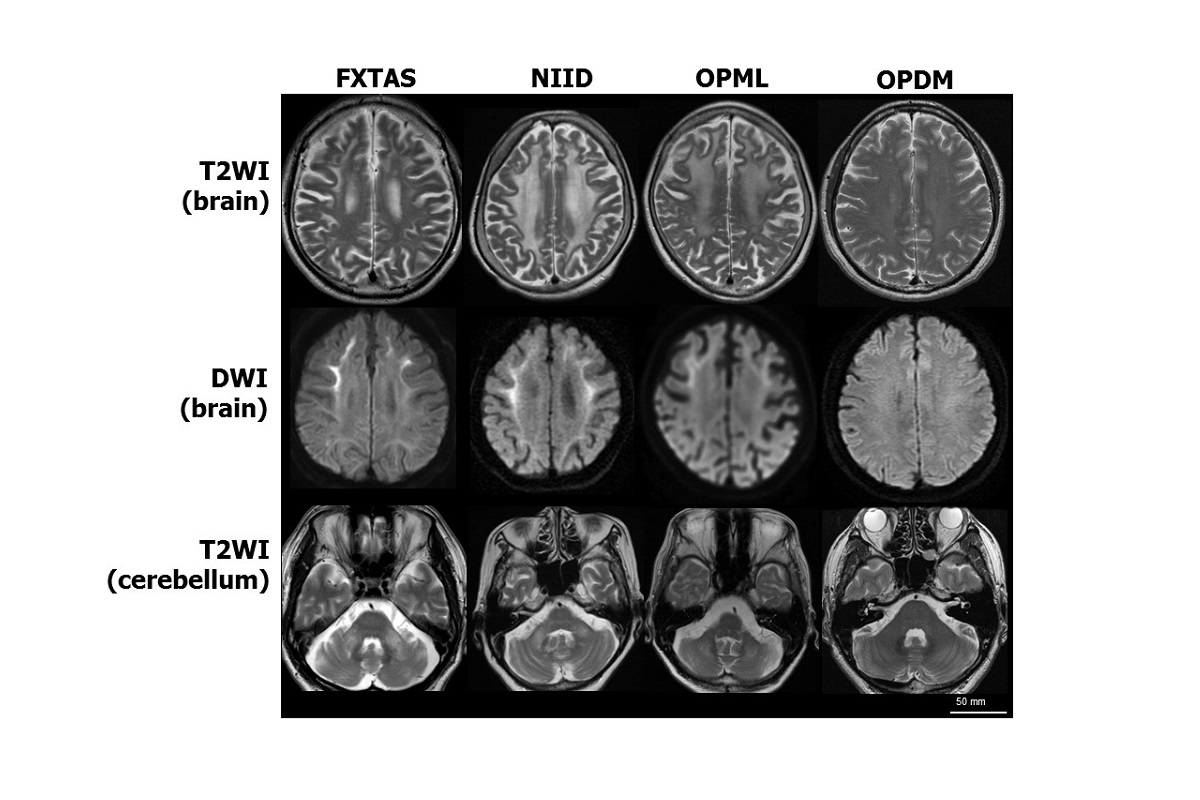
MRI brain scans of patients with similar neurodegenerative symptoms. Four rare diseases are characterized by similar symptoms of neurodegeneration. Patients with three of the diseases — fragile X tremor/ataxia syndrome (FXTAS), neuronal intranuclear inclusion disease (NIID) and oculopharyngeal myopathy with leukoencephalopathy (OPML) — have similar MRI brain scan images. Patients with a fourth disease, oculopharyngodistal myopathy (OPDM), have normal brain scans, but their muscle tissue has a similar appearance to that of patients with OPML. T2WI: T2 weighted image; DWI: Diffusion weighted image. Image by Hiroyuki Ishiura and Shoji Tsuji, CC-BY.
Families living with extremely rare neurodegenerative diseases finally received an answer to the cause of their illnesses, thanks to a researcher’s hunch and decades of improvements in DNA sequencing technology.
Four different rare diseases are all caused by the same short segment of DNA repeated too many times, a mutation researchers call noncoding expanded tandem repeats. Researchers suspect variations of this type of mutation may cause other diseases that have thus far evaded diagnosis by genetic testing.
Treatment strategies for many genetic diseases are complicated by the fact that different mutations can cause the same disease. For example, Parkinson’s disease can be caused by unique mutations in at least five different genes. Cystic fibrosis can be caused by over 1,000 different mutations in the same gene.
Researchers are excited because instead of finding unique mutations in specific genes, they identified the same mutation in different areas of the genome causing different diseases.
Genetic cause of rare diseases evaded diagnosis
“Because the mutations causing the diseases are so similar, in the future, all these patients might benefit from the same treatment,” explained Dr. Hiroyuki Ishiura, M.D./Ph.D., assistant professor from the University of Tokyo Hospital and first author of the recent research paper published in Nature Genetics.
“Gene silencing techniques [which inactivate previously active genes] are a possible treatment. We cannot know the result, but we believe such strategies may help patients in the future,” said Dr. Shoji Tsuji, M.D./Ph.D., project professor from the University of Tokyo Hospital, a corresponding author of the recent research paper.
The research team focused on patients with adult-onset neurodegeneration, showing symptoms like cognitive impairment, uncontrolled movement, loss of balance, weakness in the arms and legs, or difficulty swallowing.
The genetic cause of one disease with those symptoms, fragile X tremor/ataxia syndrome, was identified in the early 2000s as three letters of the genetic code, CGG, being repeated dozens or hundreds of times on the X chromosome. Noncoding expanded tandem repeat mutations can be caused by any letters of the genetic code repeated an unusual number of times anywhere in the genome.
Researchers had a hunch that the same CGG repeat mutation might cause three other rare diseases with similar symptoms and clinical test results. But since patients with those other diseases had normal X chromosomes, researchers had no idea where in the genome the potential CGG repeat mutations might exist.
High-tech scavenger hunt

Images from Library of Science and Medical Illustrations by somersault18:24, CC-BY-NC-SA-4.0
Previous generations of DNA sequencing technology required researchers to know where in the genome to look for a mutation. Searching for CGG repeat mutations on all 46 chromosomes in the human genome was extremely difficult and laborious.
The new approach that researchers designed relies on modern next-generation genome sequencing and clever data analysis.
Researchers sequenced patients’ and healthy people’s entire genomes in short, overlapping but broken stretches. Collaborators, including Professor Shinichi Morishita, a computational biologist from the Department of Bioinformatics and Systems Biology, developed a new computer program to sort all of those short sequences and search for ones made of just CGG over and over again.
Using a standard sequence of the entire healthy human genome, the computer program could pin down those short segments containing CGG repeat mutations to particular genetic neighborhoods. Researchers narrowed down their search to anywhere in the genome where patients had a large number of CGG repeats and healthy people had none.
With that information, researchers then knew where to sequence a long stretch of DNA to specifically identify the gene and where in the gene the patients' CGG repeat mutations exist.
Rare diseases share common symptoms
All four rare neurological diseases that the research team studied are caused by CGG repeat mutations in distant, seemingly unrelated areas of the genome.
One of the diseases is currently only known in a single family.
“We cared for this family starting 10 years ago, so it was a long puzzle for us to find the correct diagnosis,” said Ishiura.
Efforts are ongoing to study genomes identified through the genetic analysis of people who are healthy despite having the same CGG repeat mutations. Such cases are extremely rare.
The researchers hope that their studies on rare neurodegenerative diseases might lead to insights into more common diseases caused by other types of noncoding tandem repeat mutations, including amyotrophic lateral sclerosis (ALS), also known as motor neuron disease or Lou Gehrig's disease.
About the Researcher
In 2011, Tsuji received the Japanese government’s Medal with Purple Ribbon for contributions to academic development in the field of intractable neurological diseases. In 2015, he was awarded the medal for scientific achievement by the World Federation of Neurology for his leadership in the field of neurogenetics.
Papers
Ishiura H, Shibata S, Yoshimura J, et al., "Noncoding CGG repeat expansions in neuronal intranuclear inclusion disease, oculopharyngodistal myopathy and an overlapping disease," Nature Genetics: July 22, 2019, doi:10.1038/s41588-019-0458-z .
Link (Publication )
)
Related links
- Professor Shinichi Morishita’s lab

- Graduate School of Medicine

- University Hospital

- Word Federation of Neurology award announcement for Dr. Tsuji

- Medal of Honor announcement for Dr. Tsuji (Japanese)






