Understanding how cells defend their genome against invaders Researchers discover new details of pathway essential to fertility Research news

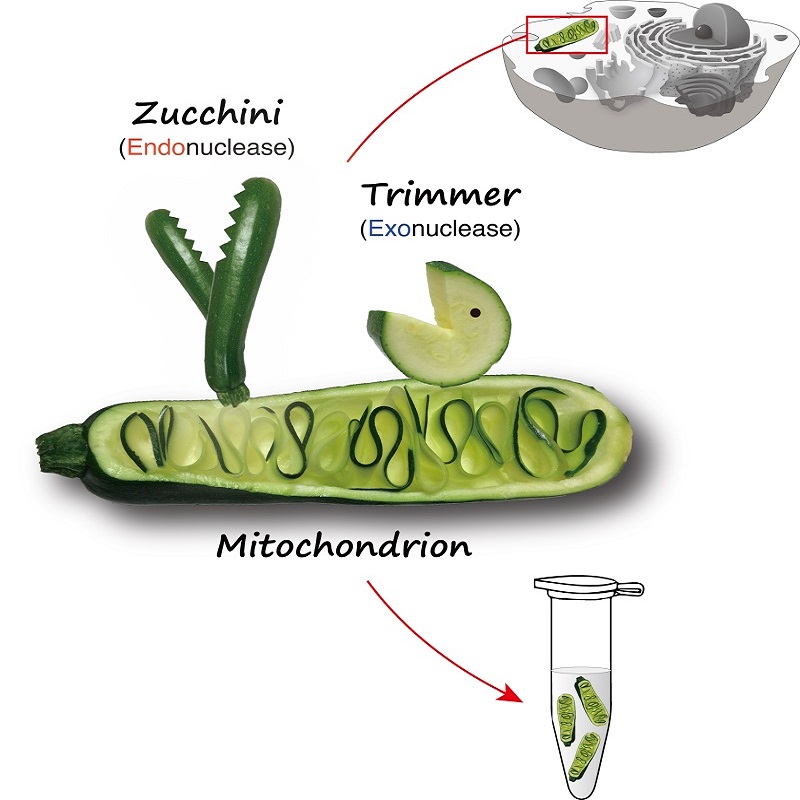
Cartoon of the portion of the cell studied in the piRNA project. © 2019 Natsuko Izumi, The University of Tokyo, CC-BY.
Researchers at the University of Tokyo have uncovered more details about the tiny defenders that ensure fertility by protecting the genomes of specialized cells called germ cells, which produce eggs and sperm. The complex system was studied using an innovative choice of research material and interdisciplinary collaboration.
The tiny genome defenders are made from short strands of RNA — DNA's molecular cousin — called piRNA.
"We are interested in how piRNAs are made because these molecules are part of a fundamental pathway that animals from insects to humans use to defend their germ cell genomes. We know that piRNA is essential for fertility and is also implicated in some cancers," said Professor Yukihide Tomari, leader of the research team at the University of Tokyo Institute for Quantitative Biosciences.
Although researchers recognize their fundamental importance, multiple challenges have made studying piRNA slow and difficult.
The UTokyo team has revealed that a protein called Zucchini processes piRNA from a long immature form into a shorter intermediate form. That intermediate piRNA is then matured into a functional form by another protein called Trimmer. Additionally, the RNA sequence that Zucchini recognizes as its signal to cut long immature piRNA is more complex than previously thought.
Pathway to piRNA
Researchers began by genetically modifying silkworm ovary cells to be unable to complete the final step of piRNA maturation by Trimmer, leaving the cells with plenty of intermediate pre-piRNA to study.
Members of the piRNA research team. From left to right: Keisuke Shoji, Yukihide Tomari, and Natsuko Izumi. © 2019 Caitlin Devor, The University of Tokyo, CC-BY.
"Obtaining the completely modified cells was a big technical challenge, not knowing what was a genuine effect of changing the piRNA pathway and what might be an accidental side effect of the genetic modification," said Research Associate Natsuko Izumi, a biochemist and co-first author of the recent publication.
Previous reports from other groups blurred the role of the Zucchini protein, a controversy Tomari attributes to the intrinsic difficulty of studying the piRNA production pathway out of the full cellular context. In fact, both Zucchini and Trimmer sit on the surface of the "energy factories" of cells, the mitochondria, but the proteins lose their original characteristics once they are purified.
The UTokyo team's innovative method of studying "crude cell pellets" where the proteins can function in a more natural situation on the mitochondrial surface revealed that indeed Zucchini does cut long immature piRNA and convert it into intermediate pre-piRNA.
Moreover, the researchers discovered how Zucchini recognizes where to cut the RNA strands.
"Next-generation sequencing can produce a huge amount of data, but as a mixture of meaningful signals and random noise. The most difficult task was to clean up the data set and organize it into a hypothesis that is experimentally testable," explained Research Associate Keisuke Shoji, a bioinformatics specialist and co-first author of the research paper.
After analysis, Shoji identified a novel motif in the RNA sequences where Zucchini prefers to cut an immature RNA strand. Additional experiments using crude cell pellets by Izumi confirmed that altering the motif prevents normal pre-piRNA production by Zucchini.
"We were surprised to know that the simple sequence signature that was believed to be the hallmark of Zucchini’s recognition is in fact not essential, but a much more complex motif is preferred by Zucchini. We imagine that the piRNA pathway is so complicated because piRNA is essential to defend the genome against diverse invader sequences and protect fertility — cells need a flexible and robust system of defense," said Tomari.
Innovations to study piRNA

Tiny molecules of piRNA play an astronomically important role in defending the germ cell genome against invading sequences; problems with piRNA lead to infertility in animals from insects to humans. This image represents two different branches of the complex pathway that produces piRNA, including the newly understood protein named Zucchini. © 2019 Natsuko Izumi, The University of Tokyo, CC-BY.
Historically, researchers have struggled to collect enough material to study the piRNA pathway because it is usually only active in germ cells.
"We have done it in the past, but dissecting ovaries out of insects is a difficult task," explained Tomari.
The first innovation came in 2009 when then-graduate student Shinpei Kawaoka and Associate Professor Susumu Katsuma at UTokyo’s Department of Agricultural and Environmental Biology detected continuous piRNA production in cells taken from the ovary of a silkworm and grown in a laboratory dish.
However, new discoveries about piRNA were still based only on genetic analysis, not direct observation of the molecules involved in producing piRNA. The normal technique of breaking cells open and analyzing the "clean" lightweight liquid portion revealed only that the piRNA pathway is not active there.
In 2011, Tomari's research team together with Kawaoka, who now leads his own research group at Kyoto University, and Katsuma made their second innovation.
"It was serendipitous," Tomari remembers.
Since nothing else was working, Kawaoka tried analyzing the portion of cells that is normally discarded in biochemistry — what Tomari refers to as the "crude, dirty pellet" made of all the solids left over after cells are broken open.
"For modern biologists this may seem like a crazy idea, but this approach is the key to our studies," said Tomari.
Then in 2016, Izumi carefully separated different components in the crude cell pellet and discovered that the piRNA processing activities reside on the mitochondrial surface, which led them to uncover the long-sought identity of the Trimmer enzyme.
Since developing these innovations, the research team has worked to define the pathway of piRNA production in detail and plans to continue characterizing the earlier and later stages of the piRNA pathway.
Papers
Natsuko Izumi, Keisuke Shoji, Yutaka Suzuki, Susumu Katsuma, and Yukihide Tomari, "Zucchini consensus motifs determine the mechanism of pre-piRNA production," Nature, doi:10.1038/s41586-020-1966-9.
Link (Publication )
)
Related links
- Lab of RNA Function (Tomari Lab)

- Department of Computational Biology and Medical Sciences, Graduate School of Frontier Sciences

- Institute for Quantitative Biosciences






