RNAPII jumps to take a shortcut Interdisciplinary team develops new model of eukaryote gene transcription

In eukaryotes, RNA polymerase II (RNAPII) is responsible for the fundamental cellular process of transcription, by which the information encoded in genes is read and encoded in a segment of RNA. However, it is difficult to mathematically describe transcription in eukaryotic cells because of the complex and organized behavior of RNAPII molecules in live cells.
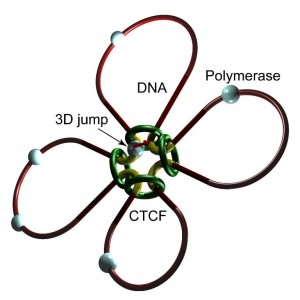
An RNAPII molecule becomes able to jump from the end of the gene (an exon) to the beginning of the next gene (next exon) with probability p, except at the edge of the last gene (last exon).
© Y Ohta and S Ihara
An interdisciplinary team lead by S. Ihara and Y. Ohta (Research Center of Advance Science and Technology), and T. Tokihiro (Graduate School of Mathematical Sciences) solved this problem by introducing a new transportation path. In the new model, just as a driver can avoid a traffic jam by taking a different route, RNAPII can make a three-dimensional jump from one site to another in addition to the usual one-dimensional motion along the DNA. This new transportation path also explains biological function in the newly discovered RNA polymerase clusters called transcription factories. The model can quantitatively provide transcription-related properties of all genes via computer simulation.
Since cellular activity depends on transcription, this method will help exploring disease pathology, accelerate new drug discovery, and may shorten time?to-market of new drugs and treatments.
Press releasePaper
Yoshihiro Ohta, Akinobu Nishiyama, Yoichiro Wada, Yijun Ruan,Tatsuhiko Kodama, Takashi Tsuboi, Tetsuji Tokihiro, and Sigeo Ihara,
“Path-preference cellular-automaton model for traffic flow through transit points and its application to the transcription process in human cells”,
Physical Review E doi: 10.1103/PhysRevE.86.021918.
Article link
Links
Research Center for Advanced Science and Technology
Laboratory for Systems Biology and Medicine
Ihara Laboratory (Japanese)






