Partial atomic structure of a eukaryote flagellar motor Structure of the microtubule-binding domain of flagellar dynein

A research group lead by Yusuke Kato and Masaru Tanokura (Department of Applied Biological Chemistry, the University of Tokyo) and Stan Burgess (Astbury Centre for Structural Molecular Biology, University of Leeds, UK) has revealed the atomic structure of the microtubule-binding domain (MTBD) of the eukaryotic flagellar motor protein, flagellar dynein.
© 2014 Yusuke Kato, Masaru Tanokura.
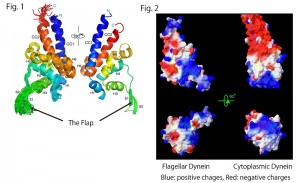
Fig. 1. Superposition of the 20 best structures of the MTBD of flagellar dynein and the nomenclatures of secondary structure elements with rainbow colors from the N- to C-termini (blue to red) (left). Mean structure viewed from the opposite side (right). The panels were depicted with Pymol (Schrodinger, LLC., New York, NY). Fig. 2. Surface representation with calculated electrostatic charges of MTBDs of flagellar dynein (left) and cytoplasmic dynein (right). The panels were depicted with Swiss PDB viewer (Guex and Peitsch, 1997).
Flagellar dyneins drive the beating motions of cilia and flagella by moving back and forth on the regularly-arranged microtubule pairs in the flagellum. Dysfunction in flagellar dyneins is associated with diverse medical conditions including male infertility and situs inversus (mirrored internal organ localization), which results from abnormalities in cell motility.
The present study has revealed that flagellar dynein MTBD possesses a novel flexible flap structure unseen in the structure of cytoplasmic dynein MTBD. Even more surprisingly, the surface charge distribution of the MTBD of flagellar dynein also differs greatly from that of cytoplasmic dynein.
In combination with biochemical analysis, these results together suggest that the mechanisms of microtubule-binding and binding affinities are different in these two dyneins. Such different properties of flagellar dynein may be important for flagellar dynein to function in a coordinated manner with numerous other dynein molecules in the confinement of the flagellum. The results obtained in the present study are important for elucidating the detailed mechanism of flagellar motion in eukaryotic organisms.
Calculated molecular dynamics of the microtubule binding domain (MTBD) of flagellar dynein.
© 2014 Sarah A Harris, Yusuke Kato, Masaru Tanokura and Stan A Burgess.
Overview of three-dimensional structure the microtubule-binding domain (MTBD) of flagellar dynein.
© 2014 Yusuke Kato, Stan A Burgess and Masaru Tanokura.
Press release (Japanese)
Paper
Yusuke S. Kato, Toshiki Yagi, Sarah A. Harris, Shin-ya Ohki, Kei Yura, Youske Shimizu, Shinya Honda, Ritsu Kamiya, Stan A. Burgess, and Masaru Tanokura,
“Structure of the microtubule-binding domain of flagellar dynein”,
Structure, Online Edition: 2014/10/24 (Japan time), doi: 110.1016/j.str.2014.08.021.
Article link
Links
Graduate School of Agricultural and Life Sciences
Department of Applied Biological Chemistry, Graduate School of Agricultural and Life Sciences
Dr. Stan Burgess, Astbury Centre for Structural Molecular Biology, University of Leeds






