Paving the way for live-cell omics Nondestructive, rapid measurement of cellular transcriptome by Raman microscopy

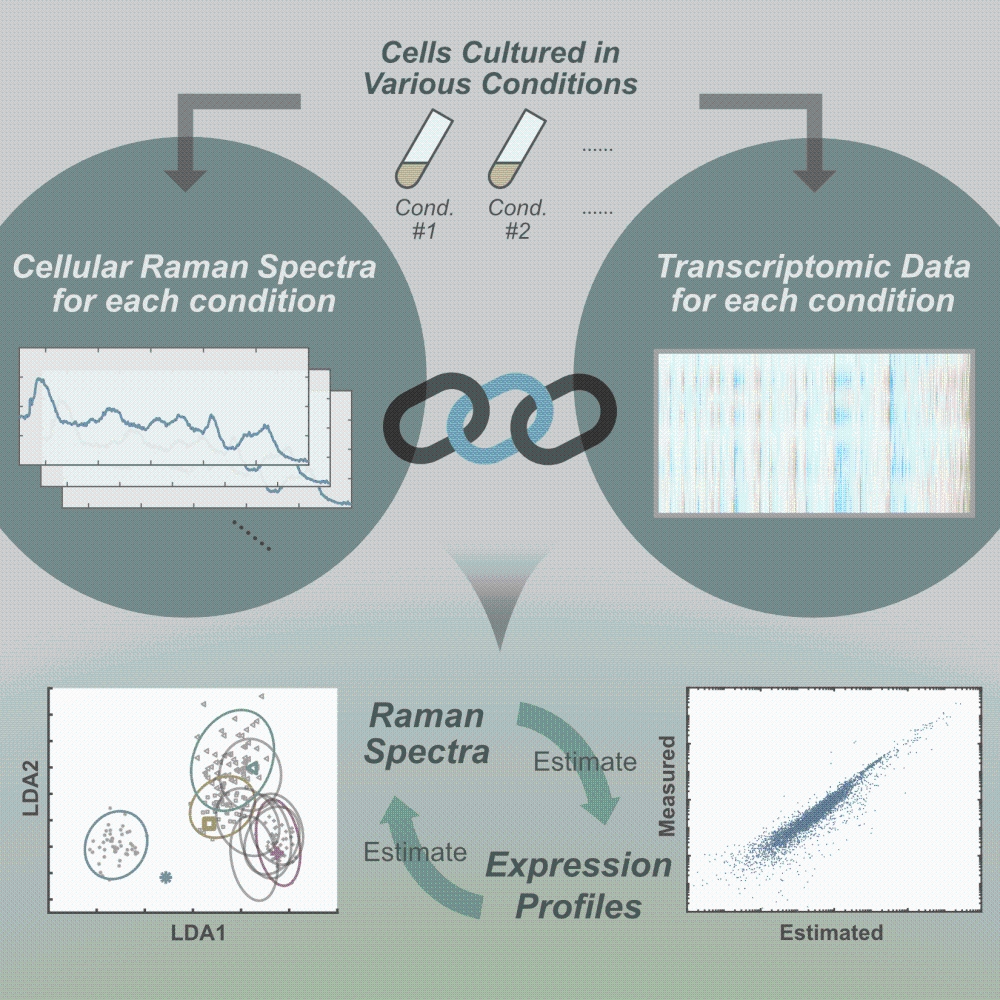
University of Tokyo researchers have unveiled the linkage between transcriptomic profile and cellular Raman spectra, which allows scientists to predict the expression levels of thousands of genes from the Raman spectra in a snapshot-like manner.
A cell contains thousands of genes, and condition-dependent expression of those genes is thought to determine the cellular state collectively. However, the conventional methods for measuring comprehensive molecular compositions of cells such as transcriptome and proteome require the destruction of cells to extract intracellular RNAs and proteins to determine the amount of each component. Therefore, the methods cannot be applied to living cells.
Raman spectroscopy is a method gaining attention that allows the measurement of signals coming from all molecules in living cells. It is a laser-based analytical technique that measures the shift in energy of scattered photons caused by molecular bond vibrations. Specific molecules have unique Raman spectral signatures, which in turn allows scientists to determine the chemical species in target samples. However, owing to the complex molecular compositions of cells, discerning constituent molecular species in comprehensive manners is widely recognized as difficult, making the interpretation of cellular Raman spectra nearly impossible.
Instead of pursuing the spectral decomposition, University of Tokyo researchers led by Associate Professor Yuichi Wakamoto and graduate student Koseki Kobayashi-Kirschvink at the Graduate School of Arts and Sciences took a different approach: They searched for the linkage between transcriptome and cellular Raman spectra computationally. They obtained Raman spectra from single cells of fission yeast, Schizosaccharomyces pombe, cultured under 10 different conditions, and found that those spectra can be classified in a low-dimensional space by a computational method called principal component linear discriminant analysis (PC-LDA). They estimated the correspondence between transcriptomes and Raman spectra by partial least square regression (PLS-R). The estimated correspondence allowed them to predict the transcriptome from cellular Raman spectra with high precision for the datasets that were excluded when estimating the regression parameter. A statistical analysis revealed that the probability of obtaining the correspondence of such high precision by chance is smaller than 0.04 percent, which offered strong support for the correspondence between transcriptomes and Raman spectra. Furthermore, they confirmed the Raman-transcriptome correspondence in Escherichia coli, a bacteria species found in animal intestines, and predicted the transcriptomes of this organism from the Raman spectra. These results additionally provided an important biological insight that the changes of transcriptomes are intrinsically low-dimensional.
Cellular Raman spectra are measurable by a few seconds of laser exposure without destructing a cell unlike conventional omics measurements. Furthermore, Raman measurements do not require the sample preparation procedure of extracting the cellular molecules such as RNAs and proteins. Therefore, the Raman-transcriptome correspondence characterized in this study leads to nondestructive, rapid and inexpensive measurement of transcriptomes, and paves the way toward conducting spectroscopic live-cell omics studies.
“The correspondence between cellular Raman spectra and transcriptomes is far from trivial because transcriptomes targeted in our study constitute only a small fraction of biomass, about 2 percent. In addition, the contribution of total RNAs to the total Raman signal is considered minor. The Raman-transcriptome correspondence was therefore unexpected and intriguing for understanding how whole-cell molecular composition is determined,” says Wakamoto. He continues, “The results in this study are important not only for the practical application to realize live-cell omics, but also for understanding how cells work; it is implied that the global expression changes of thousands of genes in cells are intrinsically low-dimensional.”
Papers
Koseki J. Kobayashi-Kirschvink, Hidenori Nakaoka, Arisa Oda, Ken-ichiro F. Kamei, Kazuki Nosho, Hiroko Fukushima, Yu Kanesaki, Shunsuke Yajima, Haruhiko Masaki, Kunihiro Ohta, Yuichi Wakamoto, "Linear Regression Links Transcriptomic Data and Cellular Raman Spectra," Cell Systems: June 20, 2018, doi:10.1016/j.cels.2018.05.015.
Link (Publication )
)
Related links
- Graduate School of Arts and Sciences

- Department of Basic Science, Graduate School of Arts and Sciences

- Wakamoto Lab, Department of Basic Science, Graduate School of Arts and Sciences






