Inference, prediction, and control of growth and evolution of cell populations


- 1.2 Data science
- 3.5 Biology
Tetsuya J. Kobayashi
Institute of Industrial Science
Professor
Self-replication and evolution are fundamental properties of living organisms but are also the causes of the development of drug-resistant bacteria and cancers. With this project, we will develop a machine learning methodology to infer, predict and control the evolution of cell populations based on quantitative data. We will also apply such methods to the problems of drug resistance.
Related links
Research collaborators
Wakamoto Laboratory, Department of Basic Science, Graduate School of Arts and Sciences, The University of Tokyo
Related publications
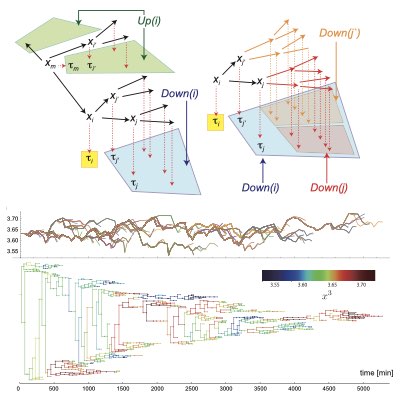
- Atsushi Kamimura and Tetsuya J. Kobayashi, Representation and inference of size control laws by neural-network-aided point processes, Phys. Rev. Research 3, 033032, 2021.7, https://researchmap.jp/7000018565/published_papers/33099937
- So Nakashima, Yuki Sughiyama, Tetsuya J Kobayashi, Lineage EM algorithm for inferring latent states from cellular lineage trees, Bioinformatics, 36, 9, 2829–2838, 2020.5, https://researchmap.jp/7000018565/published_papers/24549838
SDGs
Contact
- Tetsuya J. Kobayashi
- Email: tetsuya[at]sat.t.u-tokyo.ac.jp
※[at]=@